A glimpse of the next generation of AlphaFold
research
Progress Update: The latest Alphafold shows great improved accuracy and expands out of proteins to other biological particles, including links
Since its launch in 2020, Alphafold has revolutionized how to understand proteins and their interactions. Google DeepMind and Isomorphic Labs work together to build the foundations of the most powerful artificial intelligence model that expands out of proteins only into a full range of biologically related molecules.
Today we share an update on the progress of the next generation of Alphafold. Our last model can now generate predictions of almost all particles in the protein data bank (PDB), often reaching atomic accuracy.
It opens a new understanding and greatly improves accuracy in the categories of multiple vital particles, including links (small molecules), proteins, DNA and RNA, and those that contain post -translation modifications (PTM). These different structure types and complexes are necessary to understand the biological mechanisms inside the cell, and have been a challenge to predict high accuracy.
The capabilities and extensive performance of the model can speed up vital medical breakthroughs and achieve the following era of “digital biology” – giving new visions in the performance of pathways, genome, ulino materials, plant immunity, potential therapeutic goals, drug design mechanisms, new platforms to enable protein and mixture engines.
A series of expected structures compared to the (white) reality of our latest alphafold model.
Top of folding protein
Alphafold was a fundamental penetration of the series of series protein. Then Alphafold-Multimer expands to compounds with multiple protein chains, followed by Alphafold2.3, which improved the performance and expanded coverage of larger complexes.
In 2022, Alphafold’s brown predictions of all the indexing proteins known to science were freely available via the alphafold protein database, in partnership with the European Biomedical Institute of Embl (Embl-EBI).
To date, 1.4 million users in more than 190 countries have reached the alphafold database, and scientists all over the world have used alphafold predictions to help progress research on everything that accelerate new malaria vaccines and progress in detecting drugs to cancer to develop enzymes that consist of plastic to treat pollution.
Here we show the wonderful alphafold capabilities to predict accurate structures that go beyond protein fold, generating high -resolution predictions through bonds, proteins, nuclear acids and adjustments after the transition.
Performance via the protein compounds (A), proteins (B), nucleic acids (C), and covalent modifications (D).
Speeding the detection of the drug
Early analysis also shows that our model greatly outperforms Alphafold2.3 over some of the problems of predicting protein structure related to drug discovery, such as linking antibodies. In addition, the prediction of protein structures to recruit an incredibly valuable tool to detect drugs, as scientists can help identify and design new particles, which can become medications.
The current standard of industry is to use “laying methods” to determine reactions between bonds and proteins. These methods of laying require a solid reference protein and a proposed position to connect to its connection.
Our last model determines a new tape to predict the protein structure to recruit by outperforming the best reported methods, without the need for a reference protein structure or a jeep-recruitment site-which allows predictions of completely new proteins that have not been structurally distinguished before.
It can also design all atoms sites jointly, allowing them to represent the flexibility of proteins and nuclear acids during their interaction with other molecules – which is not possible using laying methods.
Here, for example, there are three cases that have been recently published with a therapeutic connection as the expected structures of our latest style (colorful in color) coincide closely with the experimentally specified structures (as shown in gray):
- Porcn: Anti -Cancer molecule linked to its goal, along with another protein.
- Chairs: A triple complex with a tshti recruitment (molecular glue) for an important cancer goal.
- PI5P4kγ: Selective ALLOSTERIC inhibitors of fat quinase, with multiple effects of disease including cancer and immune disorders.
Porcn predictions (1), Kras (2), and PI5P4Kγ (3).
Isomorphic Labs apps this alphafold form of the next generation to the treatment of therapeutic drugs, which helps to prescribe many types of large molecular structures and accuracy for the treatment of the disease.
A new understanding of biology
By opening protein and legends modeling with nuclear acids and those that contain post -promotional adjustments, our model provides a faster and accurate tool for studying basic biology.
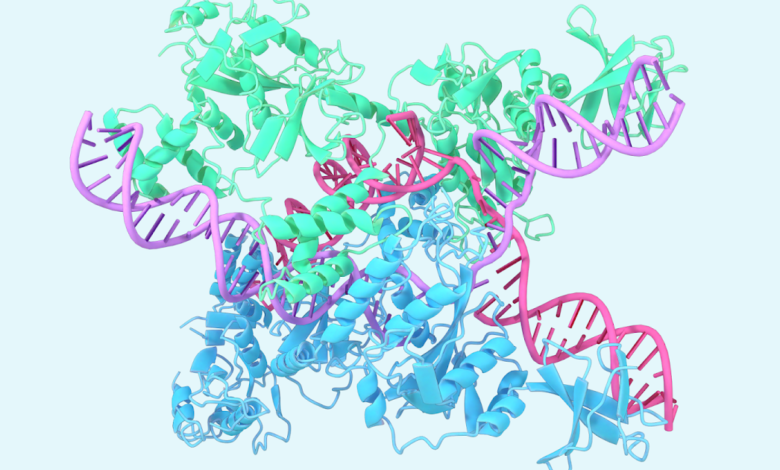
One example involves the Caslambda structure associated with CRRNA and DNA, part of the Krisper family. Caslambda shares the ability of the genome editing of CRISPR-LAS9, usually known as “genetic scissors”, which researchers can use to change the DNA of animals, plants and microorganisms. The smaller caslambda size may allow more efficient use in the mege of the genome.
An expected structure of Caslambda (CAS12L) is binding CRRNA and DNA, part of the CRISPR sub -system.
The latest version of Alphafold’s ability to design such complex systems explains to us that artificial intelligence can help us to better understand these types of mechanisms, and accelerate their use of therapeutic applications. More examples are available in updating progress.
Progress scientific exploration
Our dramatic performance in performance shows the potential of Amnesty International to enhance the scientific understanding of molecular machines that make up the human body – and the broader nature of nature.
Alphafold has already stimulated major scientific developments around the world. Now, the next generation of Alphafold has the ability to help promote scientific exploration with digital speed.
Our custom teams in Google DeepMind and Isomorphic Labs have made great steps forward about this decisive work and we look forward to our continuous progress.
2023-10-31 13:00:00




