AlphaProteo generates novel proteins for biology and health research
research
The new artificial intelligence system designs proteins that are successfully associated with targeted molecules, with the possibility of advancing medicines and understanding diseases and more.
Each biological process in the body, from cell growth to immune responses, depends on interactions between the molecules called proteins. Like a lock key, protein can be associated with another, which helps to regulate critical cellular processes. We have already gave us a protein structure like alphafold a huge vision about how proteins interact with each other to perform their functions, but these tools cannot create new proteins to process these reactions directly.
However, scientists can create new proteins successfully associated with targeted molecules. These volumes can help researchers accelerate progress through a wide range of research, including the development of medicines, cell photography, tissues, understanding of diseases and diagnosis – even crop resistance to pests. Although modern protein design methods have taken great steps, the process is still arduous and requires a wide experimental test.
Today, we offer Alphaproteo, our first Amnesty International system to design new high -strength protein folders to serve as building blocks for biological and health research. This technology has the ability to accelerate our understanding of biological processes, help in discovering new drugs, developing vital sensors and more.
Alphaproteo can generate new protein stadiums for various targeted proteins, including Vegf-A, which are associated with cancer and complications from diabetes. This is the first time that the artificial intelligence tool has been able to design a successful protein documented for VEGF-A.
Alphaproteo also achieves higher and 3 to 300 times trial success rates of binding connections than the best current methods of seven targeted proteins we tested.
Learn the complex methods that the proteins are related to each other
Protein folders that can be tightly associated with targeted protein. Traditional methods are intense of time, and require multiple tours of comprehensive laboratory work. After creating the folders, they are undergoing additional experimental tours to improve the convergence of fastening, so that it is tightly connected enough to be useful.
It was trained in large quantities of protein data from the protein data bank (PDB) and more than 100 million expected structures from Alphafold. Alphaproteo has learned that the countless molecules that are linked to the molecules with each other. Looking at the structure of a targeted molecule and a group of preferred link sites on this molecule, alphaproteo is born a filter protein associated with the goal in those sites.
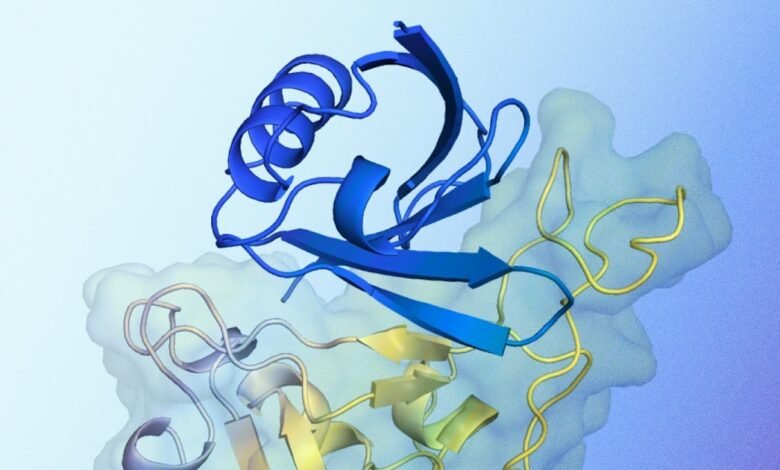
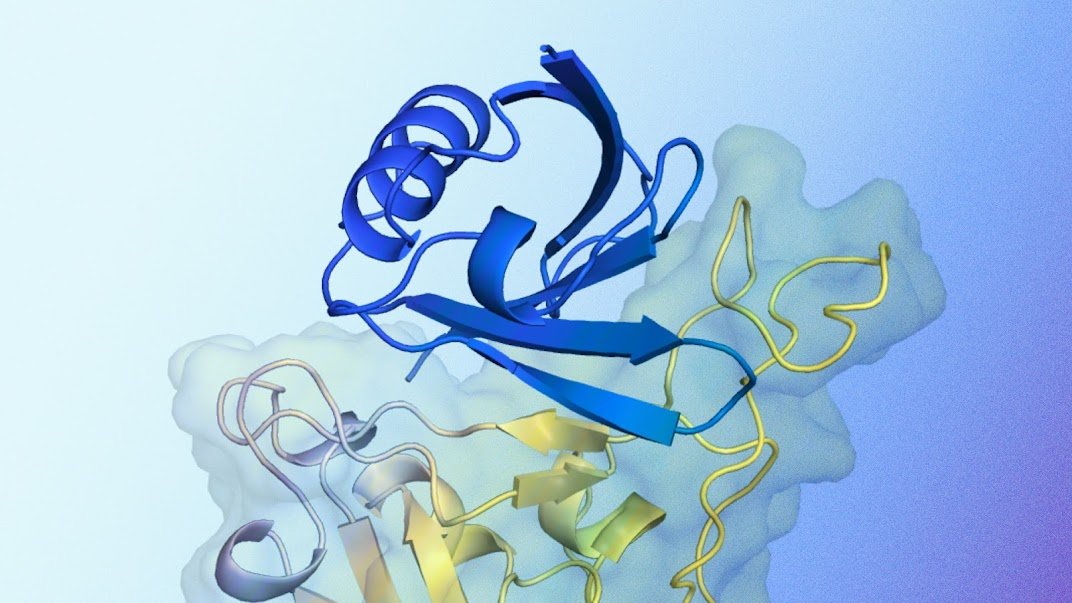
A clarification of the expected protein documented structure interacts with the target protein. Blue appears with the expected protein documented structure created by alphaproteo, designed to connect to the target protein. Yellow is the target protein, specifically the field of linking SARS-COV-2 receptors
Show success on the important protein links
To test alphaproteo, we designed the folders for various target proteins, including viral proteins involved in infection, BHRF1 and SARS-COV-2 linked to protein receptors, SC2RBD, five proteins involved in cancer, inflammation and self-bachelors, IL-7Rɑ, PD-L1,, Traka, IL-11A.
Our system contains highly competitive and binding success rates and better connection points in their class. For seven goals, Alphaproteo has created Silico -candidate proteins that are strongly associated with their intended proteins when testing them experimentally.
A network of illustrations of the expected structures of seven targeted proteins that were born Alpharoteo successful folders. Blue color appears examples of folders tested in the wet laboratory, as shown in yellow protein targets, and are distinguished in dark yellow are intended bonding areas.
For one specific goal, the BHRF1 viral protein, 88 % of our candidate particles successfully obligated when tested at the Google DeepMind Wet Laboratory. Based on the targets tested, Alphaproteo folders also link 10 times, on average, one of the best current design methods.
For another goal, TKA, our folders are stronger than the best designed folders for this goal that has gone through multiple trips of experimental improvement.
The tape graph, which appears experimentally in the success rates in the laboratory to remove Alphaproteo for each of the seven targeted proteins, compared to other design methods. Top success rates mean that less designs must be tested to find successful folders.
The tape chart that shows the best convergence of Alphaproteo designs without experimental improvement for both the seven targeted proteins, compared to other design methods. Lower rapprochement means that the documented protein is tightly linked to the target protein. Please note the logarithmic scale of the vertical axis.
Verify our results
behind In silico Check the health and test alphaproteo in our wet laboratory, we participated in the PEER CHEREPANOOV research groups, Katie Bentley and David LV Bauer from the Crikc Institute to verify the health of our protein folders. Through different experiments, they are deeper in some of the strongest SC2RBD and Vegf-A. Research groups have confirmed that binding interactions of these volumes were already similar to the predictiono. In addition, the groups confirmed that the folders have a useful biological function. For example, some of our SC2RBD folders were displayed to prevent SARS-COV-2 and some variables of infected cells.
Alphaproteo’s performance indicates that it can significantly reduce the time needed for primary experiences that involve protein programs for a wide range of applications. However, we know that our artificial intelligence system has restrictions, because it has not been able to design successful folders for the eighth goal, TNFɑ, a protein associated with autoimmune diseases such as rheumatoid arthritis. We have chosen TNFɑ to strongly challenge AlPhaproteo, as the arithmetic analysis showed that it would be very difficult to design folders against. We will continue to improve and expand the capabilities of AlPhaproteo in order to address these difficult goals in the end.
Usually, achieving a strong link is the first step only in the design of proteins that may be beneficial for practical applications, and there are many obstacles to biomedic engineering that must be overcome in the research and development process.
Towards the responsible development of protein design
Protein design is a rapidly developed technique that carries a lot of capabilities to develop science in everything from understanding the factors that cause the disease, to accelerate the development of diagnostic tests for spreading viruses, supporting the most sustainable manufacturing processes, and even cleaning pollutants from the environment.
To take into account the potential risks of biological security, and to build on our long -term approach to responsibility and safety, we are working with senior external experts to inform our teacher’s approach to sharing this work, and nutrition in society’s efforts to develop best practices, including NTI’s new biological forum.
To move forward, we will work with the scientific community to take advantage of alphaproteo on the problems of influencing biology and understand its limits. Its drug design applications have been explored in ISOMORPHIC LABS, and we are excited about the future.
At the same time, we continue to improve the success rate and the convergence of alphaproteo algorithms, expand the scope of design problems that you can deal with, and work with researchers in machine learning, structural biology, biochemistry and other specializations to develop a responsible and more comprehensive protein design for society.
If you are a scientist, his research can benefit from connecting the target protein, and you want to record interest in being a reliable test on Alphaproteo, please contact us on Alphaproteo@gooogle.com.
We will process the messages received according to privacy policy.
Thanks and appreciation
This research was developed by the protein design team and the Wet Laboratory team.
We would like to thank our collaborators Peter Chipanov, David Power and Katie Bentley and their collections at the Francis Kreik Institute for their experimental vision and invaluable results, and the Alfafold team, whose work previously contributed to this program.
2024-09-05 15:00:00