Google DeepMind Releases AlphaGenome: A Deep Learning Model that can more Comprehensively Predict the Impact of Single Variants or Mutations in DNA
A unified deep educational model for understanding the genome
Google DeepMind revealed alphagenomeA new deep educational framework designed to predict the regulatory results of the DNA sequence changes through a wide range of biological methods. Alphagenome is accepted by accepting the long DNA-1 megabyan sequence-and producing high-resolution predictions, such as basic levels of linkage, chromatin arrival, genetic expression, and linking the copy factor.
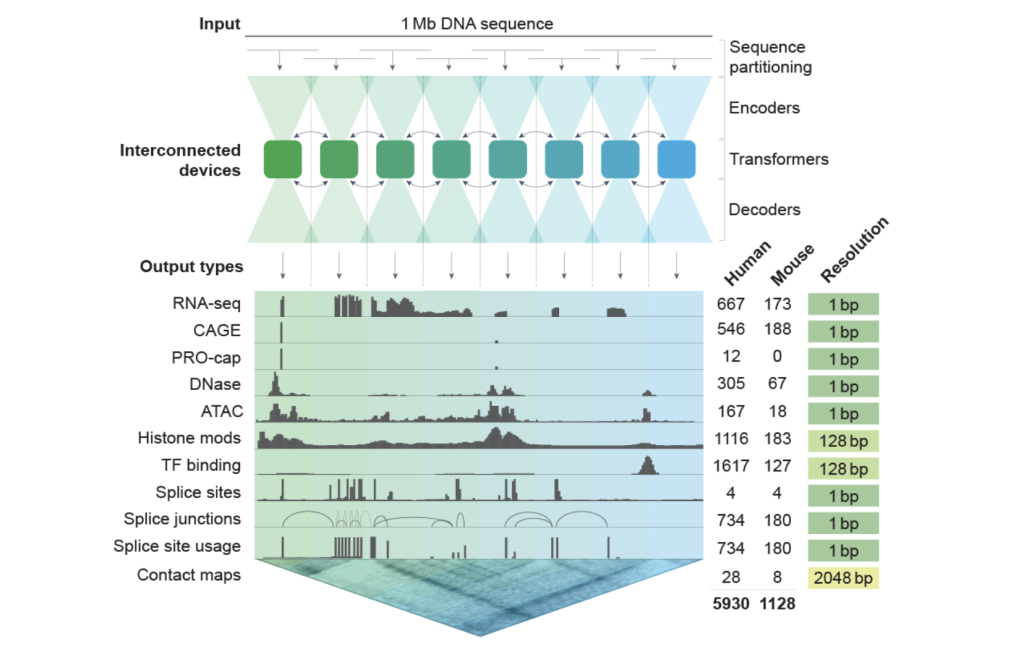
It is designed to handle restrictions in previous models, and alphagenome blocks the gap between the processing of long inputs and the resolution of the result at the level of nucleotides. The predictive tasks unite through 11 output methods and take over more than 5,000 human genetic paths and 1000+ mouse track. This level of multimedia power places alphagenome as one of the sequence models to the most comprehensive function in the genome.
Technical architecture and training methodology
Alphagenome adopts a Architecture similar to U -Net With a substance transformer. The DNA sequences in parallel parts are treated 131 km via TPUV3 devices, allowing predictions known for context and basic accuracy expectations. Architecture uses two -dimensional contents of spatial reaction modeling (for example, contact maps) and mono -dimensional functions of linear genetic tasks.
Training included two stages:
- Before training: Using folding and all aircraft models to predict the monitored experimental paths.
- distillation: The student’s model learns from the models of teachers to provide consistent and effective predictions, which allows rapid reasoning (~ 1 second for each variable) to graphics processing units such as NVIDIA H100.
Performance through standards
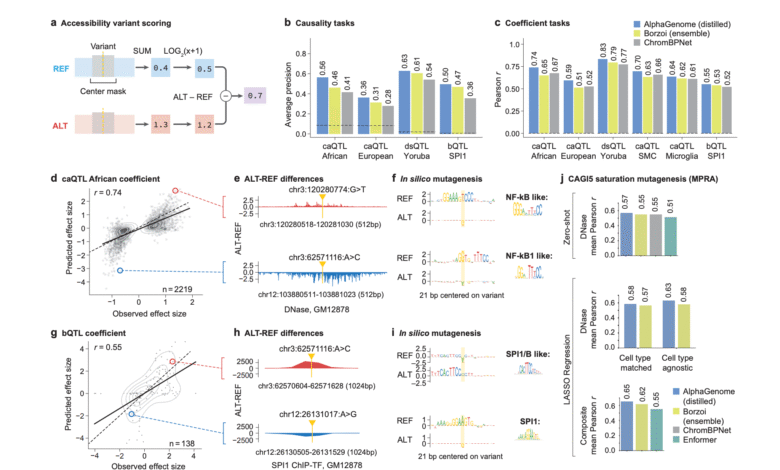
Alphagenome was strictly measured against specialized and multi -media models across 24 genome and 26 tasks to predict the effect of the variable. It excelled or identical to modern models in the 22/24 and 24/26 rating, respectively. In linking, genetic expression, and tasks related to chromatine, they have constantly exceeded specialized models such as government educational, Perzoy, and chromite.
For example:
- Bonding: Alphagenome is the first time of paste sites simultaneously, the use of paste site, and paste intersections with 1 basis point. Pangolin and SCLEAI surpassed 6 of 7 criteria.
- Eqtl predictionThe model achieved a relative improvement of 25.5 % in predicting effect compared to Borzoi.
- Access to chromatin: Show a strong association with DNASE-CEQ and ATAC-SEQ experimental data, surpassing Chrombnet by 8-19 %.
Prediction the alternative effect of the sequence alone
One of the main strengths lies in Alphagenome Predicting alternative effect (VEP). It deals with a zero deal and affects the VEP tasks without relying on population genetics, making it strong for rare variables and remote regulatory areas. With one conclusion, Alphagenome evaluates how the mutation can affect the patterns of fastening, expression levels and chromatin – all of this in a multimedia way.
The ability of the model to Clinically reproduce cluster disordersLike Exxon skipping or forming a new intersection, explaining its benefit in diagnosing rare genetic diseases. He accurately carried out 4BP deletion effects in the DLG1 gene, which was observed in GTEX samples.
The app in the interpretation of GWAS and the analysis of the disease variable
Alphagenome helps to explain GWAS signals by setting changing effects on genetic expression. Compared to Colocalization such as Coloc, Alphagenome feet supplementary and broader coverage – where the 4x sites are replaced in the lowest map.
It also showed a benefit in the genome of cancer. When analyzing non-coded mutations in the direction of the source of ONCOGENE TAL1 (associated with T-COL), Alphagenome predictions match known changes and expression expression mechanisms, confirming their ability to assess job earnings in organizational elections.
TL; D
Alphagenome by Google Deepmind is a strong, deep educational model that predicts the effects of DNA mutations through multiple regulatory methods with the accuracy of the base. It combines long -range sequence modeling, multimedia prediction, and high -resolution output in a uniform structure. Performance outperforms specialized and public models through 50 criteria. Alphagenome greatly improves the interpretation of non -encrypted genetic variables and is now available in a preview to support genome research all over the world.
verify Paper and technical details and the Japembage page. All the credit for this research goes to researchers in this project. Also, do not hesitate to follow us twitter And do not forget to join 100K+ ML Subreddit And subscribe to Our newsletter.
Asif Razzaq is the CEO of Marktechpost Media Inc .. As a pioneer and vision engineer, ASIF is committed to harnessing the potential of artificial intelligence for social goodness. His last endeavor is to launch the artificial intelligence platform, Marktechpost, which highlights its in -depth coverage of machine learning and deep learning news, which is technically sound and can be easily understood by a wide audience. The platform is proud of more than 2 million monthly views, which shows its popularity among the masses.

Don’t miss more hot News like this! Click here to discover the latest in AI news!
2025-06-26 07:39:00




