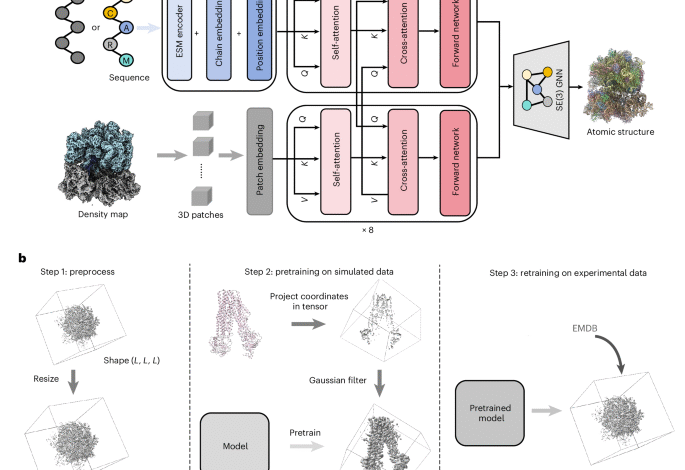
End-to-end cryo-EM complex structure determination with high accuracy and ultra-fast speed
BODU, F., Cao, H. & Cheng, J. Combining protein sequence and structures with transformers and nerve networks the graphic drawing to predict the function of protein. Biomatic informatics 39I318 – i325 (2023).
Bai, x.-C. , McMullan, G. & Siersres, SH HOW CRYO-De Revolution in structural biology. Biological trends. Sci. 4049-57 (2015).
Lawson, cl et al. EMDATARESORCE CRYO-AEM modeling results. Nat. Blind 211340-1348 (2024).
DHAKAL, A., Gyawali, R., Wang, L. & Cheng, J. Large Data collection from experts printed from experts to capture automated learning protein particles. Sci. Data 10392 (2023).
Dhakal, A., Gyawali, R., Wang, L. & Cheng, J Biomatic informatics 40BTAE109 (2024).
GIRI, N., ROY, RS & Cheng, J. Deep Learning to rebuild protein structures from cold density maps: modern developments and future trends. Curr. Opinion. structure. Biol. 79102536 (2023).
Emsley, P., Lohkamp, B., Scott, WG & Cowtan, K. Features and Development COOT. Acta Crystallogr. Sect. Diol. Crystallogr. 66486-501 (2010).
Croll, Ti Isolde: A realistic physical environment for building models in low -resolution electron maps. Acta Crystallogr. Sect. D: Brown. Biol. 74519-530 (2018).
Gao, Y., Thorn, V. & Thorn, A. Errors in structural biology are not the exception. Acta Crystallogr. Sect. The structure of Dr. Biol. 79206-211 (2023).
Croll, ti et al. Make the invisible enemy visible. Nat. structure. mall. Biol. 28404-408 (2021).
Zhong, ED, bepler, T., Berger, B. & Davis, JH Cryodrgin: Rebuilding inatistical Cryo-Es Cynics using nerve networks. Nat. Blind 18176-185 (2021).
Ranjan, R. And others. CryodrGn-ET: Reconstruction of deep obstetric networks for the perception of dynamic vital molecules inside the cells. Nat. Blind 211537-1545 (2024).
Levi, a. ADV. Nervousness. practical. split. 3513038-13049 (2022).
The scientific researcher from Google
PFab, J., PHAN, NM & SI, D. Deeptacer for Fast de Novo Cryo-Em-AM Brucky Modeling and Special Studies on COV Related Complexes. Brook. Natl Acad. Sci. USA 118Eza 2017525118 (2021).
Ronneberger, O., Fischer, P. & Brox, T in Brook. MicCai 2015: The Eighteenth International Conference, Part Three Folder. 18, 234-241 (Springer, 2015).
Hoffman, KL & Padberg, M. et al. The seller’s problem in travel. encyclopedia. a job. Accuracy. administration. Sci. 11573-1578 (2013).
The scientific researcher from Google
Jamali, K. et al. Building automatic models and determining protein in Cryo-E maps. nature 628450-457 (2024).
Rabiner, L. & Juang, B. An introduction to Markov models. IEEE AssP Mag. 34-16 (1986).
TERashi, G., Wang, X., Prasad, D., Nakamura, T Nat. Blind 21122-131 (2024).
Jumper, J. et al. Predicting with a very accurate protein structure with alphafold. nature 596583-589 (2021).
GIRI, N. & Cheng, J Nat. communication. 155511 (2024).
Vaswani, a. Attention is all you need. in Brook. Progress in nervous information processing systems Folder. 30 (Eds Guyon, I. Et Al.) 6000-6010 (Curran Associas, 2017).
Lawson, cl et al. Emdatabank data supplier for 3MM. Nuclear acids are accurate. 44D396 – D403 (2016).
Zhang, Y. & Skolnick, J. TM-ALIGN: The algorithm algorithing the protein structure on the basis of TM-SCore. Nuclear acids are accurate. 332302-2309 (2005).
Chen, C.-FR, Fan, Q. & Panda, R. Crossvit: Cross-Attenion Multi-Scale Transformer for Classification of Images. in Brook. International Conference IEEE/CVF on computer vision 357-366 (IEEE, 2021).
Satorras, VG, HogeBoom, E. & Welling, M. E (N) Equivariant Graph Neuterworks. in Brook. International Conference for Illorinary Learning 9323-9332 (PMLR, 2021).
Han, c. , Rong, J. Preprint at https://arxiv.org/abs/2202.07230 (2022).
Giri, N., Wang, L. & Cheng, J Sci. Data 11458 (2024).
Terwilliger, TC, Adams, PD, Affonine, PV & Soblev, OV is a fully automatic method that results in preliminary models of high -resolution electronic microscope maps. Nat. Blind 15905-908 (2018).
Mariani, V., BIASINI, M., Barbato, A. & Schwed, T Biomatic informatics 292722-2728 (2013).
BASU, S. & Wallner, B. DockQ: A Quality Assage for Protein Protein Models. Plos one 11E0161879 (2016).
Pintilie, G. Et al. Measurement of corn solution in the Cryo-E maps with S.-SCores. Nat. Blind 17328-334 (2020).
Van Heel, M. & Schatz, M. Fourier Shell Thresthold Thresthold. J. Struct. Biol. 151250-262 (2005).
Yamashita, K., Palmer, CM, Burnley, T Biol. Crystallogr. 771282-1291 (2021).
The scientific researcher from Google
Allegretti, M., Mills, DJ, McMullan, G., Kühlbrandt, W. & Vonck, J. Atomic Model of the F420-value [NiFe] Hydrogenease by the endoscopy of the electron using a direct electron detector. Elife 3E01963 (2014).
Bartesagi, A., Matthies, D., Bnerjee, S., MERK, A. & Subramaniam, S. β-2.2-2 3.2-obtained by CRYO electron microscope. Brook. Natl Acad. Sci. USA 11111709-11714 (2014).
Hattne, J. et al. The international radioactive damage to the location in Cryo-Im. building 26759-766 (2018).
Lynn, g. And others. Prediction of an evolutionary scale of the protein structure at the atomic level with a language model. sciences 3791123-1130 (2023).
Young, IT & Van VLIET, LJ Lying Execution of Al -Gousi candidate. Signal process. 44139-151 (1995).
Loshchilov, I. et al. Reforming weight decay in Adam. Preprint on https://arxiv.org/abs/1711.05101 (2017).
Smith, LN periodic learning rates for nervous network training. in Brook. 2017 IEEE Winter Conference on computer vision apps (WACV) 464–472 (IEEE, 2017).
Gao, Z., Tan, C. & Li, Sz Foldtoken4: Consistent and Pyramidal Language. Preprint in BIORXIV https://doi.org/10.1101/2024.08.04.606514 (2024).
Ingraham, JB et al. Lighting protein space with a programmed obstetric model. nature 6231070-1078 (2023).
Chombooon, K., Chujai, P., Teerassamee, P., KERDPRASOP, K. & Kerdprrasop, N. Experimental study of distance standards for yourA neighbor algorithm. in Brook. The third international conference on industrial application engineering Folder. 2, p. 4 (Industrial Applications Institute, 2015).
Giri, N., Wang, L. & Cheng, J. Cryo2structdata: Full Data set. Harvard Dateverse https://doi.org/10.7910/dvn/fcdg0W (2023).
Giri, N., Wang, L. & Cheng, J. Cryo2structdata: Data set test. Harvard Dateverse https://doi.org/10.7910/dvn/2GSSC9 (2023).
Wang, J. Cryfold_source_data.zip. Figshare https://doi.org/10.6084/m9.figshare.28530359.v1 (2025).
Wang, J. & Tan, C. Cryo-De Complex Contract Contracted with high accuracy and high-speed speed. Zenudo https://doi.org/10.5281/zenodo.14970359 (2025).
Hodson, to the root average square error (RMSE) or means absolute error (MAE): when to use it or not. Geosci. Dave Model. 155481-5487 (2022).
Don’t miss more hot News like this! Click here to discover the latest in AI news!
2025-06-24 00:00:00