Scalable and robust DNA-based storage via coding theory and deep learning
Rydning, Drjgj, Reinsel, J. & GANTZ, J. Drinking the world from edge to basic (international data company2018).
Meiser, LC et al. Artificial DNA applications in IT. Nat. communication. 13352 (2022).
Coze, L., Nivala, J Nat. Gennett priest. 20456-466 (2019).
Leprosy, em et al. High quality libraries of long algonokoliotides (150 Mir) through a process of controlling a new removal process. Nuclear acids are accurate. 382522-2540 (2010).
HECKEL, R., Mikutis, G. & Grass, RN Description of the DNA storage channel. Sci. representative. 99663 (2019).
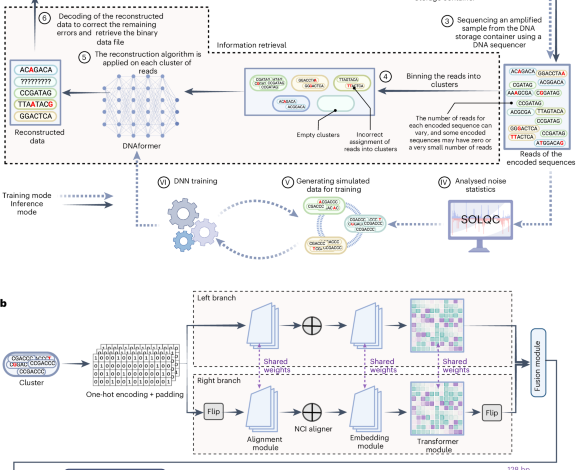
Sabari, O., Yucovich, A., Shapira, G. & Yaakobi, E. RecONSTRUCTIONTHMS for DNA storage systems. Sci. representative. 14951 (2024).
Srinivasavaradhan, SR, Gopi, S., PFister, HD & Yekhanin, S. Trellis BMA: Reintegration of encrypted tracking on IDS channels for storing DNA. in 2021 International Symposium IEEE on Information theory (ISIT) (ED. Dey, B.) 2453-2458 (IEEE, 2021); class=”c-article-references__item js-c-reading-companion-references-item” data-counter=”8.”>
Linz, a. And others. Serial recovery symbols from multiple readings for the DNA sequence. in 2020 Information theory workshop (ITW) (ED. Dalai, M.) 1-5 (IEEE, 2021).
Levenshtein, VI effective rebuilding sequences from serials or separators. J. comb. theory. progress. A 93310-32 (2001).
McGRGOR, A., PRICE, E. & Vorotnikova, S. Trace RecONSTRUCTIONIONIOND. in The European symposium on algorithms (Eds Schulz, AS & Wagner, D.) 689–700 (Springer, 2014).
Church, GM, GAO, Y. & Kosuri, S. Generation Digital Information Storage in DNA. sciences 3371628-1628 (2012).
Goldman, n. And others. Towards storing practical and high capacity information, low maintenance in the made DNA. nature 49477-80 (2013).
Grass, RN, HECKEL, R., Puddu, M., Paunescu, D. & Stark, WJ keep the strong chemistry of digital information on DNA in silica with error correction codes. Ango. Chemical. int. Mr. Dr. 542552-2555, (2015).
The scientific researcher from Google
Macwilliams, FJ & Sloane, NJA Mistake correction symbols theory Folder. 16 (Embassy, 1997).
Erlich, Y. & Zielinski, DNA Fountain allows a strong and effective storage intention. sciences 355950-954 (2017).
Organick, L. et al. Random arrival in storing DNA data widely. Nat. Biotechnology. 36242-248 (2018).
Yazdi, SMHT, Gabrys, R. & Milenkovic, O. Portable data storage and error -free. Sci. representative. 75011 (2017).
Wang, J. And others. Storage of high -capacity DNA data with changing algonoklotides length using a repetitive accumulation and hybrid maps. C. Biol. Engineer. 131-11 (2019).
Chandak, S. Et al. Overcoming high nano error rates in storing DNA by integration and entertainment symbols. in ICASSP 2020 IEEE IEEEEEENEANATIONCE on Acoustics and Cleant and Signaling (ICASSP) (ED. Pérez-Neira, AI) 8822-8826 (IEEE, 2020).
Anavy, L., Vaknin, I. ATAR, O., Ameit, R. & Yakhini, Z. DNA storage with less synthesis courses using composite DNA letters. Nat. Biotechnology. 371237 (2019).
Cherghchi, M. & Ribeiro, J. Overview of the Intestination Results of Snights Channels. IEEE Trans. Inf. theory 673207-3232 (2020).
QU, G., Yan, Z. & Wu, H Short. The vital form. 23BBAC336 (2022).
Rashtchian, C. Et al. Collect billions of readings to store DNA data. ADV. Nervousness. practical. split. 303362-3373 (2017).
Viswanathan, K. & Swaminathan, R. Reintegration of the series on the listing channels – Delete. in Brook. The nineteenth annual SIAM SIAM on the separate algorithms (ED. Teng, S.-H.) 399-408 (Industrial and Applied Mathematics Association, 2008).
BATU, T., Kannan, S., Khanna, S. & MCGREGOR, A. Rebuilding chains from random effects. Soda 4910-918 (2004).
Holden, n. in Learning theory conference Folder. 75 (Eds Bubeck, S. Et Al.) 1799-1840 (PMLR, 2018).
Holenstein, T., Mitzenmacher, M., Panigrahy, R. & Wieder, U. Trace RecONSTRUCTION with the possibility of constant deletion and relevant results. in Brook. The nineteenth annual SIAM SIAM on the separate algorithms (ED. Teng, S.-H.) 389-398 (Industrial and Applied Mathematics Association, 2008).
Nazarov, F. & Peres, Y. Trace RecONSTRUCTION with Exp (S.((N1/3)) Samples. in Brook. Annual ACM SIGACT Seminar 49 on Computing theory (Eds Hatami, H. & MCKENZIE, P.) 1042-1046 (ACM, 2017).
Peres, Y. & Zhai, A. Reintepse of the middle state of the deletion channel: many effects of many effects are sufficient. in 2017 annual symposium 58 IEEE on the foundations of computer science (FOCS) (ED. UMANS, C.) 228-239 (IEEEEEEECUTETY, 2017).
Bee, C. Et al. Search for content -based similarities in wide DNA storage systems. Nat. communication. 124764 (2021).
Pan, C. Et al. Store data -based two -dimensional DNA while rebuilding machine learning. Nat. communication. 132984 (2022).
Wolf, J. on the codes that can be derived from the tensioner product from the examination matrices. IEEE Trans. Inf. theory 11281-284 (1965).
Sabari, O. Et al. Solqc: The Oligo Synthetic Library quality control tool. Biomatic informatics 37720-722 (2021).
Preserving our digital heritage: Introduction to storing DNA data (DNA storage alliance, 2021).
GoPalan, PS et al. Reconstruction of a tumultuous polluleucleotide sequence. American patent application 15/536115 (2018).
Marcus, BH, Roth, RM, & Siegel, PH bound systems and coding for registration channels. in Introduction to coding for restricted systems (Eds Pless, V. & Huffman, WC) (Elsevier, 1998).
Gimpel, Al, Stark, WJ, HECKEL, R. & Grass, RN A Digital Twin to store DNA data based on the comprehensive amount of errors and biases. Nat. communication. 146026 (2023).
Bohlin, J., Rose, B. Big data anal. 41-11 (2019).
WeINDEL, F., GIMPEL, AL, Grass, RN & HECKEL, R. More effective embrace errors than to avoid them restricted to storing DNA data. in 2023 59 annual Allerton Conference for Communications, Control and Computing 1-8 (IEEE, 2023).
Stoler, N. & Nekrutnko, A. The Embroidery of the Illumina Sequence tools. Genome fire. The vital form. 3LQab019 (2021).
Ping, z. et al. Towards archiving practical and powerful DNA-based data using a Yin-Yang coding system. Nat. account. Sci. 2234-242 (2022).
Chaaykin, G., Foreman, N. Sabari, or. in The thirteenth annual workshop of non -volatile memories (2022).
Xiao, T. And others. Early notes help transformers a better vision. ADV. Nervousness. practical. split. 3430392-30400 (2021).
The scientific researcher from Google
Chowdhury, B. & Garai, G. A review of a multiple sequence from the genetic algorithm perspective. Genome 109419-431 (2017).
Cholete, F. XCeption: Deep Learning with DEPTHWISE accessories. in Brook. IEEE conference on computer vision and patterns 1251-1258 (IEEE, 2017).
Vaswani, a. And others. Attention is all you need. ADV. Nervousness. practical. split. 30(2017).
Menin, JF & Nichols, NM PCR MULTIPEX using high -resolution DNA polymery (New England Piolis, 2013).
Zhang, J., Kobert, K., Flouri, T Biomatic informatics 30614-620 (2014).
Wang, Y., Zhao, Y., Bollas, A., Wang, Y. & AU, KF Nanopore Sequenceing Technology, Bioinformatics and Applications. Nat. Biotechnology. 391348-1365 (2021).
Bar-Lev, D., ORR, I., Sabari, O., Etzion, T Zenudo (2024).
Bar-Lev, D., ORR, I., Sabari, O., Etzion, T Zenudo (2024).
Grass, RN, HECKEL, R., Puddu, M., Paunescu, D. & Stark, WJ for “maintaining strong chemical information for digital information on DNA in silica with error correction symbols.” Zenudo (2015).
The grass, RN and others. Data set for “maintaining strong chemical information for digital information on DNA in silica with error correction symbols”. Zenudo (2015).
Bar-Lev, D., ORR, I., Sabari, O., Etzion, T Zenudo (2024).
Bornholt, J. et al. DNA -based archive storage system. in Brook. The twenty -first international conference for architectural support is programming and operating systems 637-649 (ACM, 2016).
Blawat, M. Et al. Correcting errors forward to store DNA data. Comput procedures. Sci. 801011-1022 (2016).
Wagner, Ra & Fischer, MJ Series Correction problem. J. ACM 21168-173 (1974).
2025-02-21 00:00:00