Repurposing Protein Folding Models for Generation with Latent Diffusion – The Berkeley Artificial Intelligence Research Blog
Plaid is a multimedia model generated simultaneously the 1D protein sequence and 3D, by learning the inherent space for protein folding models.
The 2024 Nobel Prize for Alphafold2 represents an important moment of recognition of the role of artificial intelligence in biology. What comes after that after folding the protein?
in EngravedWe are developing a method that learns a sample of the inherent area of protein folding models to Generate New proteins. You can accept The formative function and the organismAnd it can be Train on sequence databasesWhich are 2-4 requests greater than the structure databases. Unlike many gym models of the previous protein structure, PLAID processes the preparation of the problem of the joint multimedia generation: simultaneously generating both separate sequences and completely continuous structural coordinates.
From predicting the structure to drug design in the real world
Although modern works clarify the promise of the ability to spread to the creation of proteins, there are restrictions on previous models that make them inappropriate for real applications, such as:
- All generation: Many current gym models produce only spine atoms. To produce all-aatom structures and put Sidechain atoms, we need to know the sequence. This creates a multimedia generation problem that requires simultaneous generation of separate and continuous methods.
- The privacy of the organismBiology proteins for human use. You need to be manTo avoid destruction by the human immune system.
- Control specificationsDiscovering drugs and placing it in the hands of patients is a complex process. How can we define these complex restrictions? For example, even after processing biology, it may be decided that tablets are easier to transport them from the bottles, adding a new restriction to the attempt.
Birth of “useful” proteins
Simply generate proteins is not like Control The generation to get useful Proteins. How can it look like this?

For inspiration, let’s think about how to control the generation of images with symbolic text claims (example from Liu et al., 2022).
In engraved, we reflect this interface for Control specifications. The ultimate goal is to completely control the generation via a text interface, but here we consider syntactic restrictions for lovers as evidence of the concept: job and The organism:
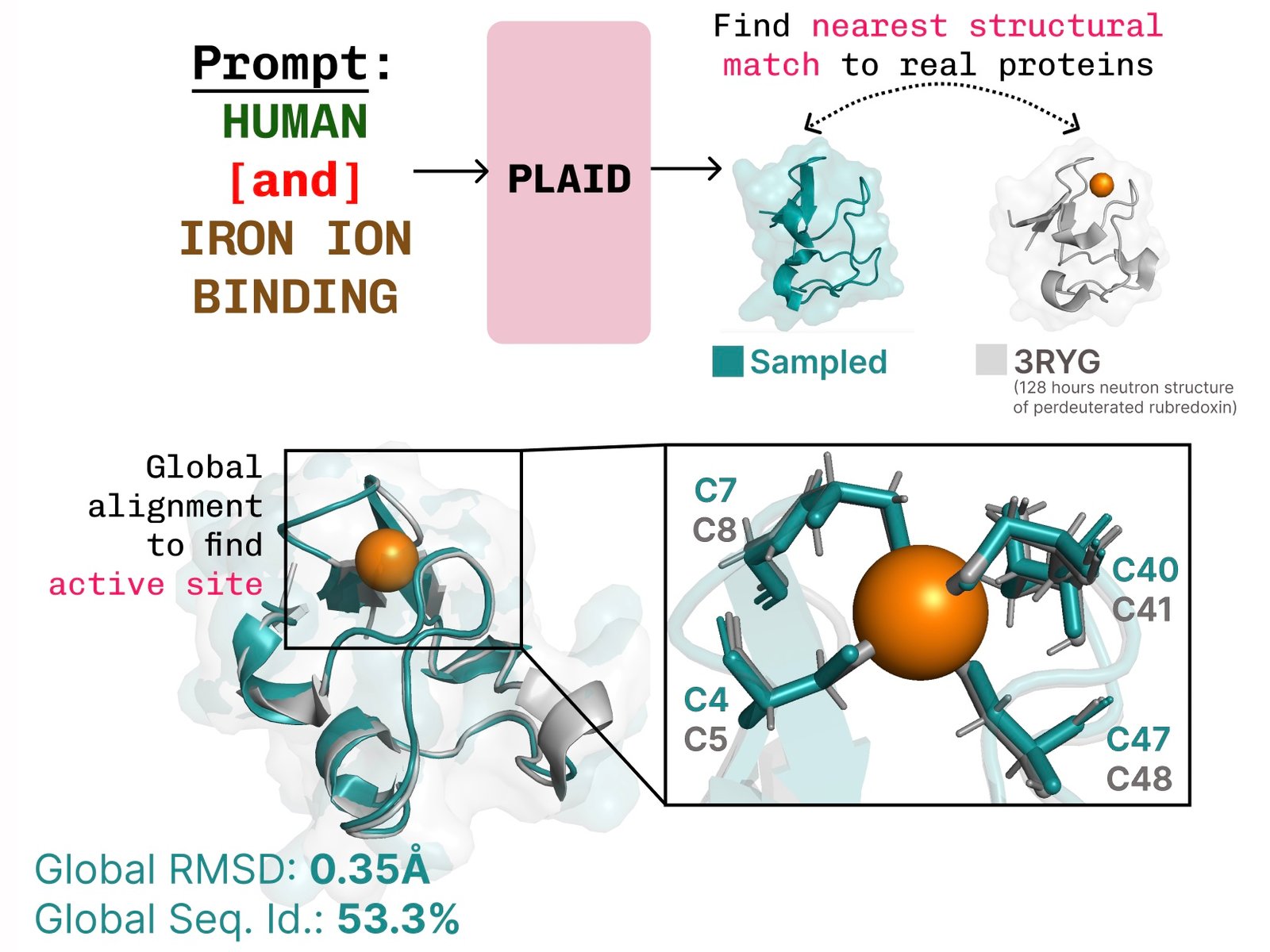
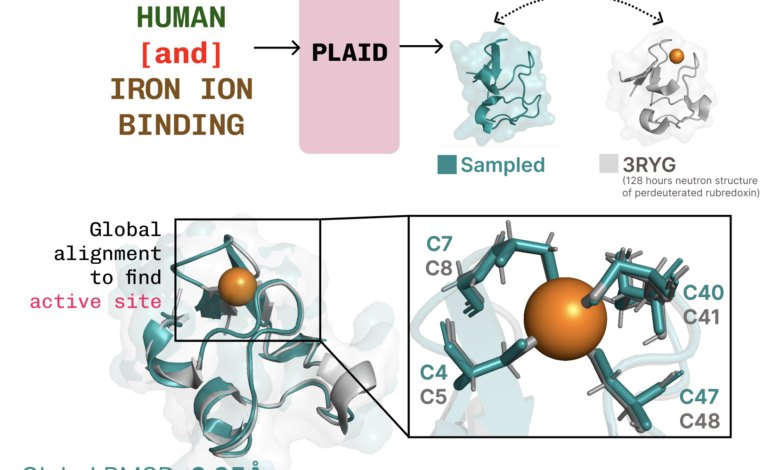
Learn to connect a job structure. Plaid learns Tetradral Cysteine-FE2+/Fe3+ The formatting pattern found in mineral proteins is often, while maintaining diversity at the high sequence level.
Training using sequence training data only
Another important aspect of the engraved model is that we just need a sequence to train the obstetric model! Gynecological models learn to distribute the specified data through their training data, and the rules of sequence data are much larger than that structure, because the sequences are much cheaper than obtaining the experimental structure.
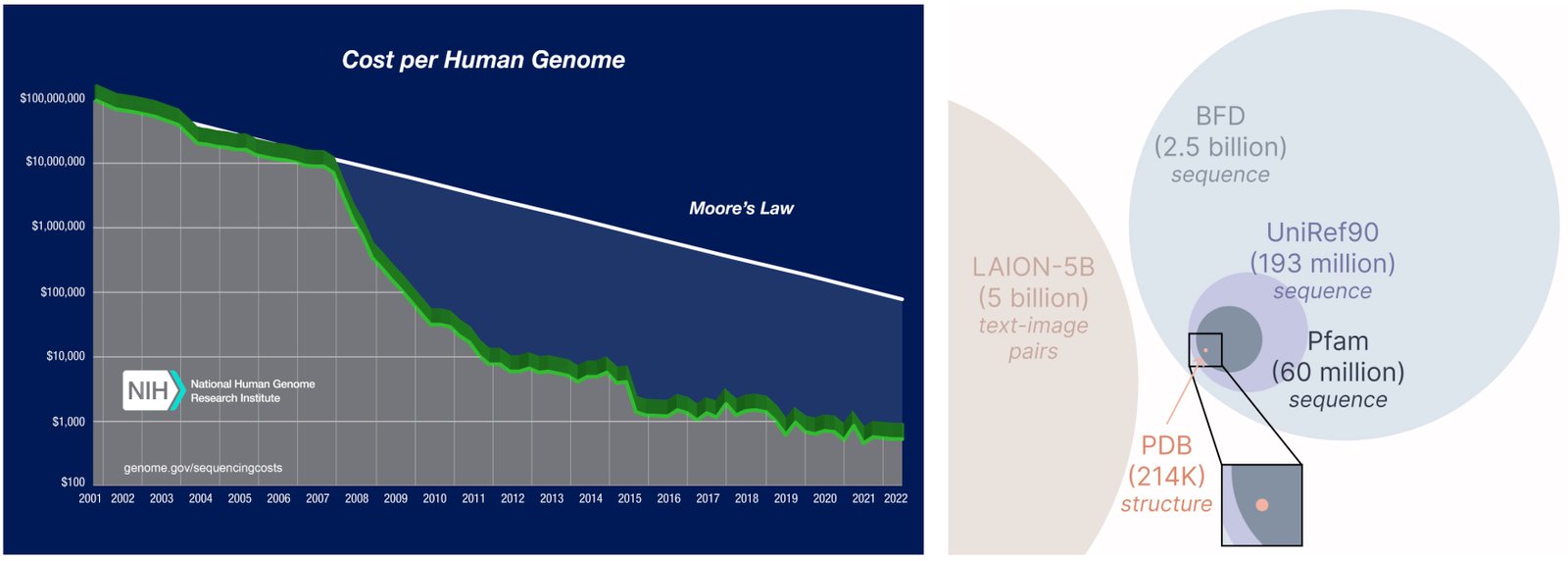
Learning from a larger and wider database. The cost of obtaining a protein sequence is much lower than the experimental structure description, and the rules of the sequence data are 2-4 orders with a size larger than the structure.
How do you work?
The reason is that we are able to train the obstetric model to create a structure by using sequence data only by learning a format on Clear space for a protein folding model. Then, while inference, after taking the samples from this potential area of good proteins, we can take Frozen From the protein folding model to brown drying. Here, we use EsMfold, a successor to the alphafold2 model that replaces a retrieval step with the protein language model.
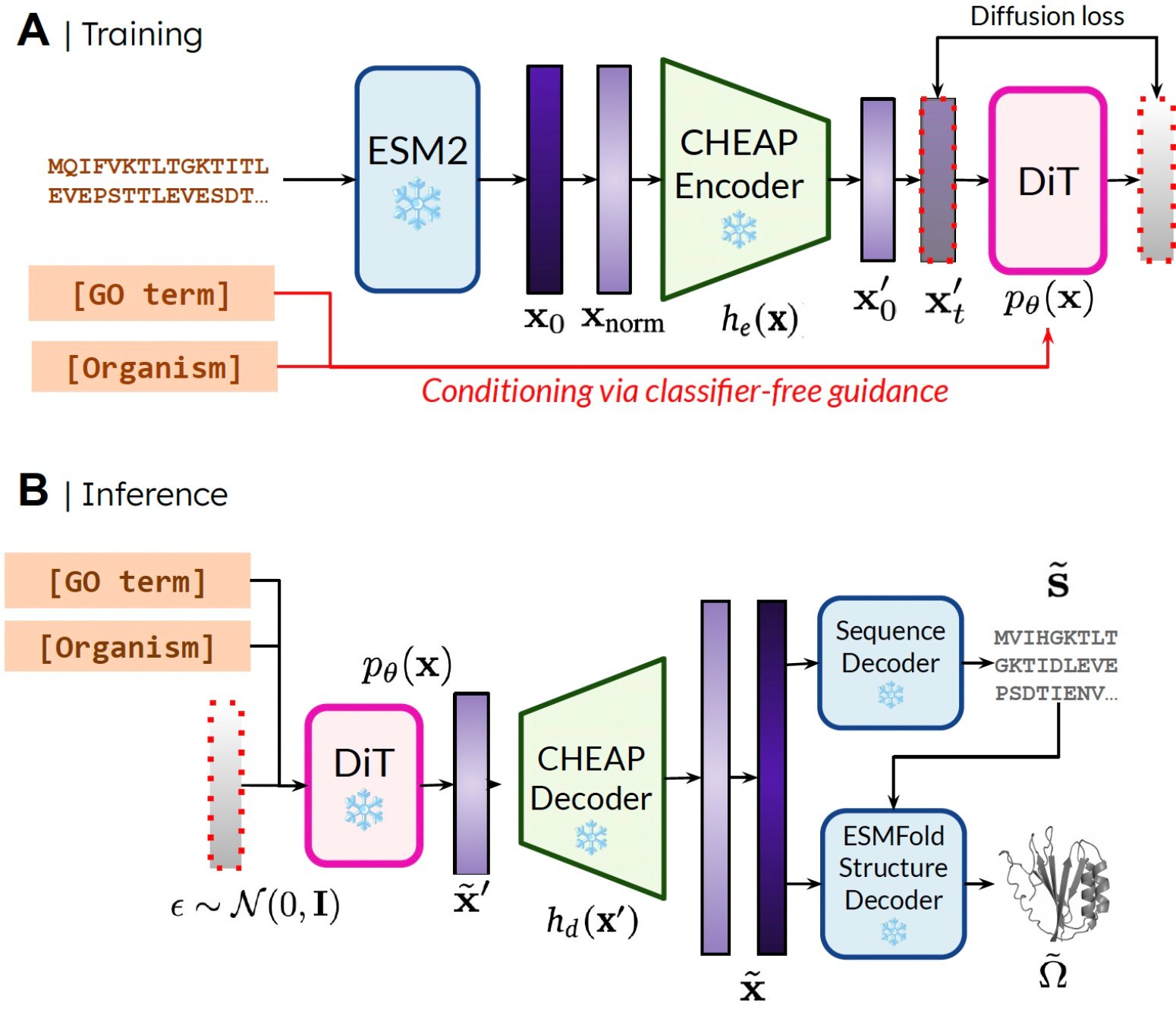
Our way. During training, only serials are needed to get inclusion; During inference, we can decipher the sequence and structure from the inclusion from which samples were taken. ❄ indicates frozen weights.
In this way, we can use structural understanding information in the weights of pre -protein folding models for the task of designing protein. This is similar to how to use language models (VLA) in robots, which were used in the vision language models trained on internet data to provide awareness, understanding and understanding information.
Compress the inherent space for protein folding models
Small wrinkle with the application of this method directly is that the potential space for ESMFOLD-in fact, the inherent space for many models based on transformers-requires a lot of organization. This space is too large, so learning this inclusion ends the maps drawing to the creation of high -resolution images.
To address this, we also suggest Cheap (Hourglass compressed including protein adaptations)Where we learn a pressure model to include the joint of the protein and structure sequence.
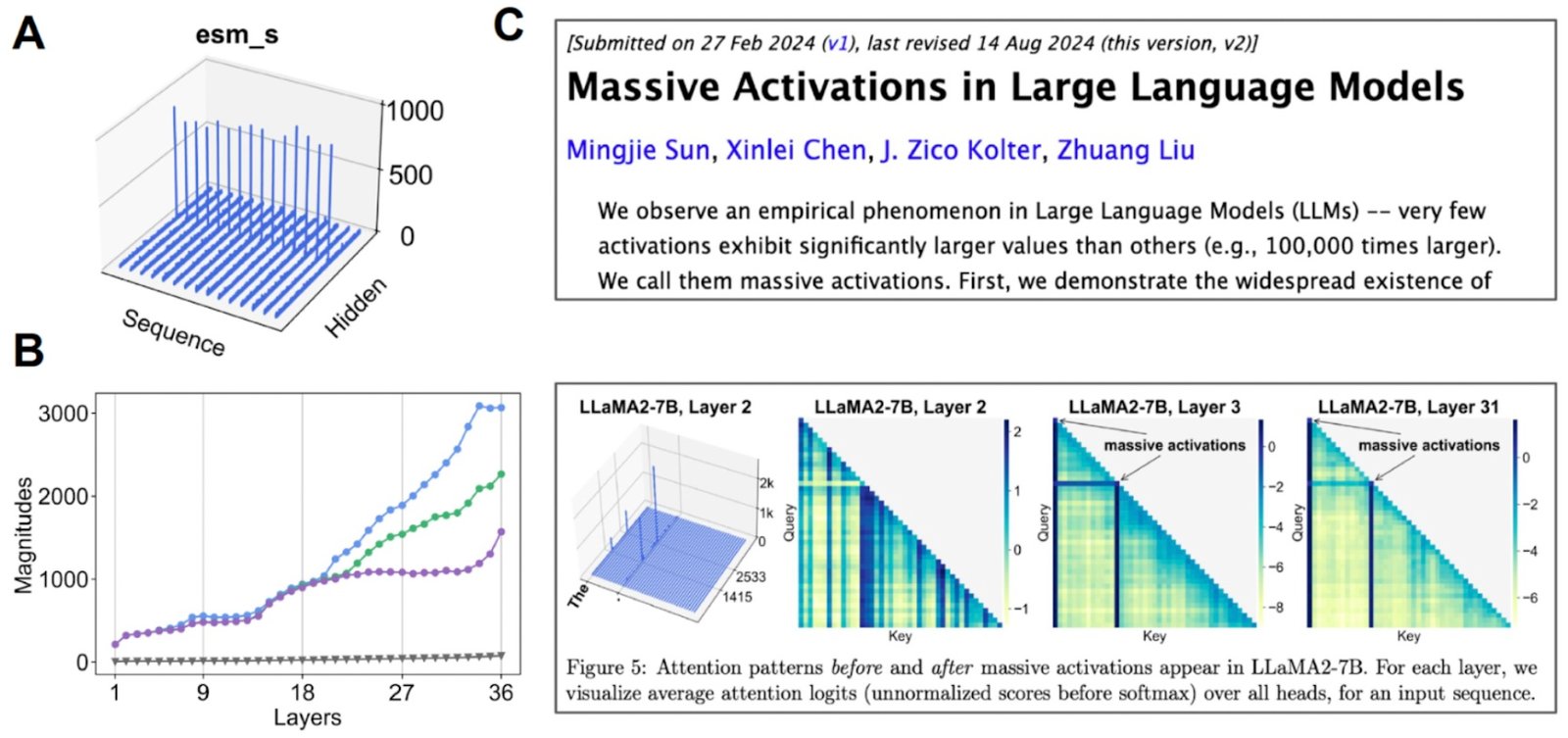
Investigate inherent space. (A) When we imagine the average value of each channel, some channels appear “huge activations”. (B) If we start examining the higher activations 3 compared to the medium (gray) value, then we find that this happens on many layers. (C) Huge activation is also observed for other transformer -based models.
We find that this inherent space is actually pressure. By doing some mechanical interpretation to understand the basic model with which we work better, we have been able to create a full protein generation model.
What next?
Although we are studying the state of protein sequence and the generation of the structure in this work, we can adapt this method to perform multimedia generation of any methods as there is a prediction of a more abundant way to a less abundant way. Since the sequence predictions to the structure of proteins began to process the increasingly complex systems (for example, Alphafold3 is also able to predict proteins in complex with nuclear acids and molecular bonds), it is easy to imagine multimedia generation performance on more complicated systems using the same method. If you are interested in cooperating to expand our way, or to test our way of wet, please contact!
Other links
If you find our papers useful in your search, please think about using the following BIBTEX for Plaid and Cheap:
@article{lu2024generating,
title={Generating All-Atom Protein Structure from Sequence-Only Training Data},
author={Lu, Amy X and Yan, Wilson and Robinson, Sarah A and Yang, Kevin K and Gligorijevic, Vladimir and Cho, Kyunghyun and Bonneau, Richard and Abbeel, Pieter and Frey, Nathan},
journal={bioRxiv},
pages={2024--12},
year={2024},
publisher={Cold Spring Harbor Laboratory}
}
@article{lu2024tokenized,
title={Tokenized and Continuous Embedding Compressions of Protein Sequence and Structure},
author={Lu, Amy X and Yan, Wilson and Yang, Kevin K and Gligorijevic, Vladimir and Cho, Kyunghyun and Abbeel, Pieter and Bonneau, Richard and Frey, Nathan},
journal={bioRxiv},
pages={2024--08},
year={2024},
publisher={Cold Spring Harbor Laboratory}
}
You can also get out of pre -prints (engraved, cheap) and codebases (engraved, cheap).
Some protein bonus fun generation!
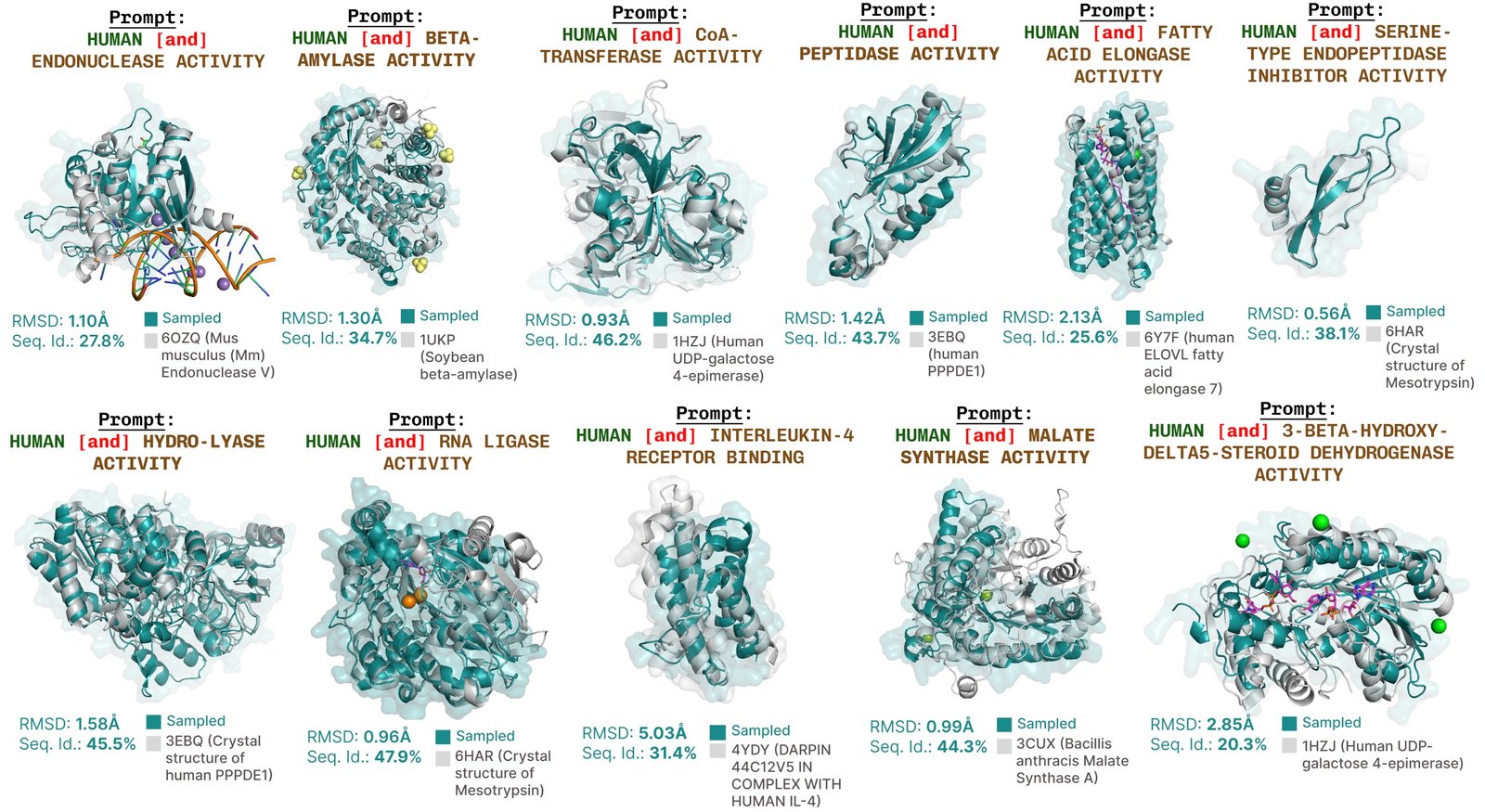
Additional generations loaded with functional with engraved.
Unlawful generation with engraved.
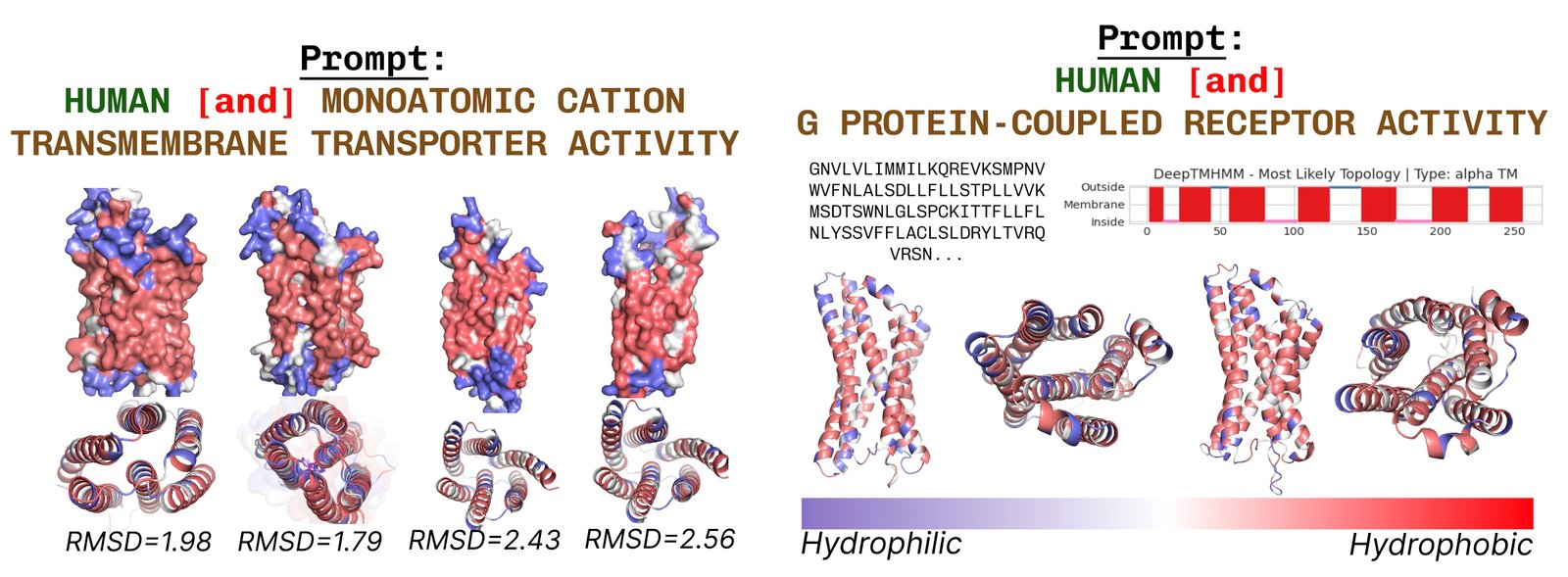
The membrane proteins contain the remains of the heart, where they are included in the fatty acid layer. These are constantly noted when demanding the cover with the protein keywords via the membrane.
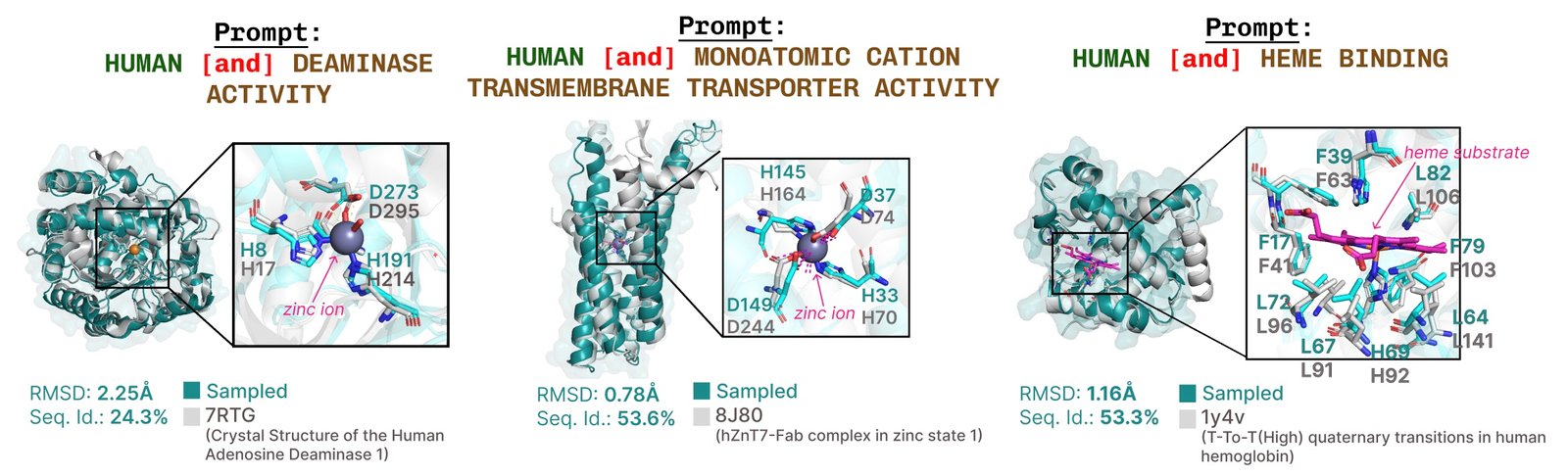
Additional examples of re -summarizing the active site based on the function of the function of the job.
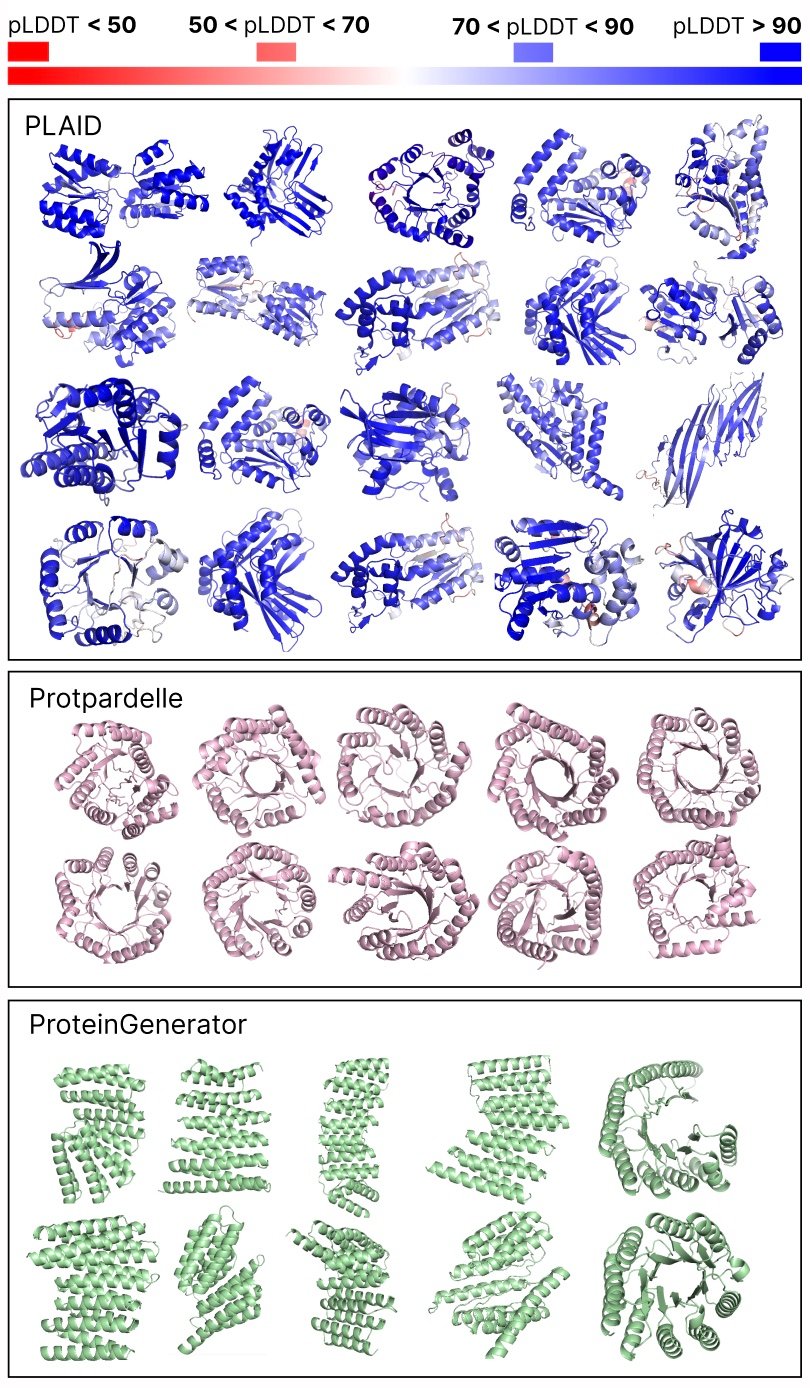
Comparison of samples between the foundation lines is engraved and all. Samples engraved have a better diversity and pick up the beta -Strand style, which was more difficult for protein generation models.
Thanks and appreciation
Thanks to Nathan Frey to detailed reactions about this article, participants around Bair, Genentech, Microsoft Research and New York University: Wilson Yan, Sarah A. Robinson, Simon Kilo, Kevin K. Yang, Vladimir Glegorigevich, Kyungi Chu, Richard Bono, Peter.
2025-04-08 10:30:00